1.2 Elements of gene regulation
The mechanisms regulating gene expression are essential for all living organisms as they dictate where and how much of a gene product (it may be protein or ncRNA) should be manufactured. This regulation could occur at the pre- and co-transcriptional level by controlling how many transcripts should be produced and/or which version of the transcript should be produced by regulating splicing. The same gene could encode for different versions of the same protein via splicing regulation.This process defines which parts of the gene will go into the final mRNA that will code for the protein variant. In addition, gene products can be regulated post-transcriptionally where certain molecules bind to RNA and mark them for degradation even before they can be used in protein production.
Gene regulation drives cellular differentiation; a process during which different tissues and cell types are produced. It also helps cells maintain differentiated states of cells/tissues. As a result of this process, at the final stage of differentiation, different kinds of cells maintain different expression profiles, although they contain the same genetic material. As mentioned above, there are two main types of regulation and next we will provide information on those.
1.2.1 Transcriptional regulation
The rate of transcription initiation is the primary regulatory element in gene expression regulation. The rate is controlled by core promoter elements as well as distant-acting regulatory elements such as enhancers. On top of that, processes like histone modifications and/or DNA methylation have a crucial regulatory impact on transcription. If a region is not accessible for the transcriptional machinery, e.g. in the case where the chromatin structure is compacted due to the presence of specific histone modifications, or if the promoter DNA is methylated, transcription may not start at all. Last but not least, gene activity is also controlled post-transcriptionally by ncRNAs such as microRNAs (miRNAs), as well as by cell signaling, resulting in protein modification or altered protein-protein interactions.
1.2.1.1 Regulation by transcription factors through regulatory regions
Transcription factors are proteins that recognize a specific DNA motif to bind on a regulatory region and regulate the transcription rate of the gene associated with that regulatory region (see Figure 1.5 for an illustration). These factors bind to a variety of regulatory regions summarized in Figure 1.5, and their concerted action controls the transcription rate. Apart from their binding preference, their concentration, and the availability of synergistic or competing transcription factors will also affect the transcription rate.
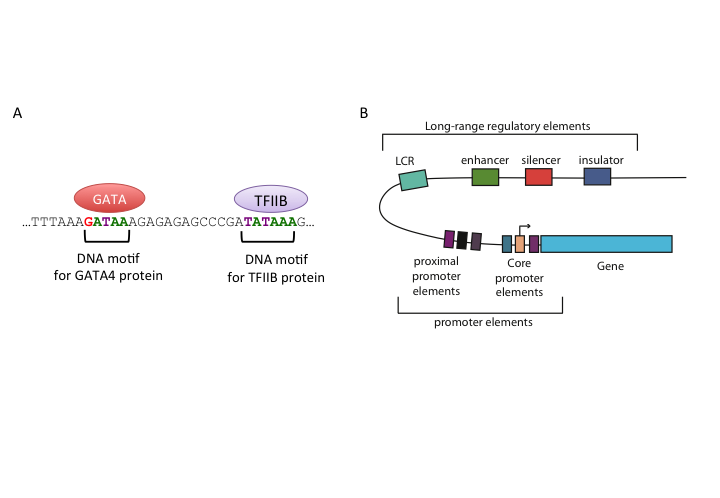
FIGURE 1.5: Representation of regulatory regions in animal genomes
1.2.1.1.1 Core and proximal promoters
Core promoters are the immediate neighboring regions around the transcription start site (TSS) that serve as a docking site for the transcriptional machinery and pre-initiation complex (PIC) assembly. The textbook model for transcription initiation is as follows: The core promoter has a TATA motif (referred as TATA-box) 30 bp upstream of an initiator sequence (Inr), which also contains TSS. Firstly, transcription factor TFIID binds to the TATA-box. Next, general transcription factors are recruited and transcription is initiated on the initiator sequence. Apart from the TATA-box and Inr, there are a number of sequence elements on the animal core promoters that are associated with transcription initiation and PIC assembly, such as downstream promoter elements (DPEs), the BRE elements and CpG islands. DPEs are found 28-32 bp downstream of the TSS in TATA-less promoters of Drosophila melanogaster. They generally co-occur with the Inr element, and are thought to have a similar function to the TATA-box. The BRE element is recognized by the TFIIB protein and lie upstream of the TATA-box. CpG islands are CG dinucleotide-enriched segments of vertebrate genomes, despite the general depletion of CG dinucleotides in those genomes. 50 to 70% of promoters in the human genome are associated with CpG islands.
Proximal promoter elements are typically right upstream of the core promoters, usually contain binding sites for activator transcription factors, and provide additional control over gene expression.
1.2.1.1.2 Enhancers
Proximal regulation is not the only or the most important mode of gene regulation. Most of the transcription factor binding sites in the human genome are found in intergenic regions or in introns. This indicates the widespread usage of distal regulatory elements in animal genomes. On a molecular function level, enhancers are similar to proximal promoters; they contain binding sites for the same transcriptional activators and they basically enhance the gene expression. However, they are often highly modular and several of them can affect the same promoter at the same time or in different time-points or tissues. In addition, their activity is independent of their orientation and their distance to the promoter they interact with. A number of studies showed that enhancers can act upon their target genes over several kilobases away. According to a popular model, enhancers achieve this by looping the DNA and coming into contact with their target genes.
1.2.1.1.3 Silencers
Silencers are similar to enhancers; however their effect is opposite of enhancers on the transcription of the target gene, and results in decreasing their level of transcription. They contain binding sites for repressive transcription factors. Repressor transcription factors can either block the binding of an activator , directly compete for the same binding site, or induce a repressive chromatin state in which no activator binding is possible. Silencer effects, similar to those of enhancers, are independent of orientation and distance to target genes. In contradiction to this general view, in Drosophila there are two types of silencers, long-range and short-range. Short-range silencers are close to promoters and long-range silencers can silence multiple promoters or enhancers over kilobases away. Like enhancers, silencers bound by repressors may also induce changes in DNA structure by looping and creating higher-order structures. One class of such repressor proteins, which is thought to initiate higher-order structures by looping, is Polycomb group proteins (PcGs).
1.2.1.1.4 Insulators
Insulator regions limit the effect of other regulatory elements to certain chromosomal boundaries; in other words, they create regulatory domains untainted by the regulatory elements in regions outside that domain. Insulators can block enhancer-promoter communication and/or prevent spreading of repressive chromatin domains. In vertebrates and insects, some of the well-studied insulators are bound by CTCF (CCCTC-binding factor). Genome-wide studies from different mammalian tissues confirm that CTCF binding is largely invariant of cell type, and CTCF motif locations are conserved in vertebrates. At present, there are two models that explain the insulator function; the most prevalent model claims insulators create physically separate domains by modifying chromosome structure. This is thought to be achieved by CTCF-driven chromatin looping and recent evidence shows that CTCF can induce a higher-order chromosome structure through creating loops of chromatins. According to the second model, an insulator-bound activator cannot bind an enhancer; thus enhancer-blocking activity is achieved and insulators can also recruit an active histone domain, creating an active domain for enhancers to function.
1.2.1.1.5 Locus control regions
Locus control regions (LCRs) are clusters of different regulatory elements that control an entire set of genes on a locus. LCRs help genes achieve their temporal and/or tissue-specific expression programs. LCRs may be composed of multiple cis-regulatory elements, such as insulators and enhancers, and they act upon their targets even from long distances. However, LCRs function in an orientation-dependent manner, for example the activity of beta-globin LCR is lost if inverted. The mechanism of LCR function otherwise seems similar to other long-range regulators described above. The evidence is mounting in the direction of a model where DNA-looping creates a chromosomal structure in which target genes are clustered together, which seems to be essential for maintaining an open chromatin domain.
1.2.1.2 Epigenetic regulation
Epigenetics in biology usually refers to constructions (chromatin structure, DNA methylation, etc.) other than DNA sequence that influence gene regulation. In essence, epigenetic regulation is the regulation of DNA packing and structure, the consequence of which is gene expression regulation. A typical example is that DNA packing inside the nucleus can directly influence gene expression by creating accessible regions for transcription factors to bind. There are two main mechanisms in epigenetic regulation: i) DNA modifications and ii) histone modifications. Below, we will introduce these two mechanisms.
1.2.1.2.1 DNA modifications such as methylation
DNA methylation is usually associated with gene silencing. DNA methyltransferase enzyme catalyzes the addition of a methyl group to cytosine of CpG dinucleotides (while in mammals the addition of methyl group is largely restricted to CpG dinucleotides, methylation can occur in other bases as well). This covalent modification either interferes with transcription factor binding on the region, or methyl-CpG binding proteins induce the spread of repressive chromatin domains, thus the gene is silenced if its promoter has methylated CG dinucleotides. DNA methylation usually occurs in repeat sequences to repress transposable elements. These elements, when active, can jump around and insert them to random parts of the genome, potentially disrupting the genomic functions.
DNA methylation is also related to a key core and proximal promoter element: CpG islands. CpG islands are usually unmethylated, however, for some genes, CpG island methylation accompanies their silenced expression. For example, during X-chromosome inactivation, many CpG islands are heavily methylated and the associated genes are silenced. In addition, in embryonic stem cell differentiation, pluripotency-associated genes are silenced due to DNA methylation. Apart from methylation, there are other kinds of DNA modifications present in mammalian genomes, such as hydroxy-methylation and formylcytosine. These are other modifications under current research that are either intermediate or stable modifications with distinct functional associations. There are at least a dozen distinct DNA modifications observed when we look across all studied species (Sood, Viner, and Hoffman 2019).
1.2.1.2.2 Histone modifications
Histones are proteins that constitute a nucleosome. In eukaryotes, eight histone proteins are wrapped by DNA and make up the nucleosome. They help super-coiling of DNA and inducing high-order structure called chromatin. In chromatin, DNA is either densely packed (called heterochromatin or closed chromatin), or it is loosely packed (called euchromatin or open chromatin). Heterochromatin is thought to harbor inactive genes since DNA is densely packed and transcriptional machinery cannot access it. On the other hand, euchromatin is more accessible for transcriptional machinery and might therefore harbor active genes. Histones have long and unstructured N-terminal tails which can be covalently modified. The most studied modifications include acetylation, methylation and phosphorylation (Strahl and Allis 2000). Using their tails, histones interact with neighboring nucleosomes and the modifications on the tail affect the nucleosomes’ affinity to bind DNA and therefore influence DNA packaging around nucleosomes. Different modifications on histones are used in different combinations to program the activity of the genes during differentiation. Histone modifications have a distinct nomenclature, for example: H3K4me3 means the lysine (K) on the 4th position of histone H3 is tri-methylated.
| Modifications | Effect |
|---|---|
| H3K9ac | Active promoters and enhancers |
| H3K14ac | Active transcription |
| H3K4me3/me2/me1 | Active promoters and enhancers, |
| H3K4me1 and H3K27ac is enhancer-specific | |
| H3K27ac | H3K27ac is enhancer-specific |
| H3K36me3 | Active transcribed regions |
| H3K27me3/me2/me1 | Silent promoters |
| H3K9me3/me2/me1 | Silent promoters |
Histone modifications are associated with a number of different transcription-related conditions; some of them are summarized in Table 1.1. Histone modifications can indicate where the regulatory regions are and they can also indicate activity of the genes. From a gene regulatory perspective, maybe the most important modifications are the ones associated with enhancers and promoters.
Furthermore, certain proteins can influence chromatin structure by interacting with histones. Some of these proteins, like those of the Polycomb Group (PcG) and CTCF, are discussed above in the insulators and silencer sections. In vertebrates and insects, PcGs are responsible for maintaining the silent state of developmental genes, and trithorax group proteins (trxG) for maintaining their active state (Henikoff 2008 ; Schwartz and Pirrotta 2007). PcGs and trxGs induce repressed or active states by catalyzing histone modifications or DNA methylation. Both the proteins bind PREs that can be on promoters or several kilobases away. Another protein that induces histone modifications is CTCF. CTCF is associated with boundaries between active and repressive histone marks (Phillips and Corces 2009). This is due to the role of CTCF in regulating the 3D genome structure. Two CTCF binding sites that are far away from each other in linear distance can bind together in 3D space thus forming chromatin loops.
Want to know more?
Transcriptional regulatory elements in the human genome: http://www.ncbi.nlm.nih.gov/pubmed/16719718
On metazoan promoters: Types and transcriptional properties: http://www.ncbi.nlm.nih.gov/pubmed/22392219
General principles of regulatory sequence function: http://www.nature.com/nrg/journal/v15/n7/abs/nrg3684.html
DNA methylation: Roles in mammalian development: http://www.nature.com/doifinder/10.1038/nrg3354
Histone modifications and organization of the genome: http://www.nature.com/nrg/journal/v12/n1/full/nrg2905.html
DNA methylation and histone modifications are linked: http://www.nature.com/nrg/journal/v10/n5/abs/nrg2540.html
1.2.2 Post-transcriptional regulation
1.2.2.1 Regulation by non-coding RNAs
Recent years have witnessed an explosion in non-coding RNA (ncRNA)-related research. Many publications implicated ncRNAs as important regulatory elements. Plants and animals produce many different types of ncRNAs such as long non-coding RNAs (lncRNAs), small interferring RNAs (siRNAs), microRNAs (miRNAs), promoter-associated RNAs (PARs) and small nucleolar RNAs (snoRNAs) (Morris and Mattick 2014). lncRNAs are typically >200-bp long, they are involved in epigenetic regulation by interacting with chromatin remodeling factors and they function in gene regulation. siRNAs are short double-stranded RNAs which are involved in gene regulation and transposon control; they silence their target genes by cooperating with Argonaute proteins. miRNAs are short single-stranded RNA molecules that interact with their target genes by using their complementary sequence and mark them for quicker degradation. PARs may regulate gene expression as well: they are approximately 18-to -200-bp-long ncRNAs originating from promoters of coding genes (Morris and Mattick 2014). snoRNAs are also shown to play roles in gene regulation, although they are mostly believed to guide ribosomal RNA modifications (Morris and Mattick 2014).
1.2.2.2 Splicing regulation
Splicing is regulated by regulatory elements on the pre-mRNA and proteins binding to those elements. Regulatory elements are categorized as splicing enhancers and repressors. They can be located either in exons or introns. Depending on their activity and their locations there are four types of regulatory elements for splicing:
- exonic splicing enhancers (ESEs)
- exonic splicing silencers (ESSs)
- intronic splicing enhancers (ISEs)
- intronic splicing silencers (ISSs).
The majority of splicing repressors are heterogeneous nuclear ribonucleoproteins (hnRNPs). If splicing repressor protein bind silencer elements, they reduce the chance of a nearby site being used as a splice junction. On the contrary, splicing enhancers are sites to which splicing activator proteins bind and binding on that region increases the probability that a nearby site will be used as a splice junction (Wang and Burge 2008). Most of the activator proteins that bind to splicing enhancers are members of the SR protein family. Such proteins can recognize specific RNA recognition motifs. By regulating splicing exons can be skipped or included, which creates protein diversity (Wang and Burge 2008).
Want to know more?
On miRNAs, their genesis, and modes of regulation (Bartel 2004): http://www.sciencedirect.com/science/article/pii/S0092867404000455
Functions of non-coding RNAs (Morris and Mattick 2014): http://www.nature.com/nrg/journal/v15/n6/abs/nrg3722.html
On splicing and its regulation (Wang and Burge 2008): https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/18369186/
References
Bartel. 2004. “MicroRNAs: Genomics, Biogenesis, Mechanism, and Function.” Cell 116 (2): 281–97.
Henikoff. 2008. “Nucleosome Destabilization in the Epigenetic Regulation of Gene Expression.” Nature Reviews Genetics 9 (1): 15–26.
Morris, and Mattick. 2014. “The Rise of Regulatory Rna.” Nature Reviews Genetics 15 (6): 423–37.
Phillips, and Corces. 2009. “CTCF: Master Weaver of the Genome.” Cell 137 (7): 1194–1211.
Schwartz, and Pirrotta. 2007. “Polycomb Silencing Mechanisms and the Management of Genomic Programmes.” Nature Reviews Genetics 8 (1): 9–22.
Sood, Viner, and Hoffman. 2019. “DNAmod: The Dna Modification Database.” Journal of Cheminformatics 11 (1): 30.
Strahl, and Allis. 2000. “The Language of Covalent Histone Modifications.” Nature 403 (6765): 41–45.
Wang, and Burge. 2008. “Splicing Regulation: From a Parts List of Regulatory Elements to an Integrated Splicing Code.” Rna 14 (5): 802–13.