Chapter 6 Operations on Genomic Intervals and Genome Arithmetic
Considerable time in computational genomics is spent on overlapping different features of the genome. Each feature can be represented with a genomic interval within the chromosomal coordinate system. In addition, each interval can carry different sorts of information. An interval may for instance represent exon coordinates or a transcription factor binding site. On the other hand, you can have base-pair resolution, continuous scores over the genome such as read coverage, or scores that could be associated with only certain bases such as in the case of CpG methylation (see Figure 6.1 ). Typically, you will need to overlap intervals of interest with other features of the genome, again represented as intervals. For example, you may want to overlap transcription factor binding sites with CpG islands or promoters to quantify what percentage of binding sites overlap with your regions of interest. Overlapping mapped reads from high-throughput sequencing experiments with genomic features such as exons, promoters, and enhancers can also be classified as operations on genomic intervals. You can think of a million other ways that involve overlapping two sets of different features on the genome. This chapter aims to show how to do analysis involving operations on genomic intervals.
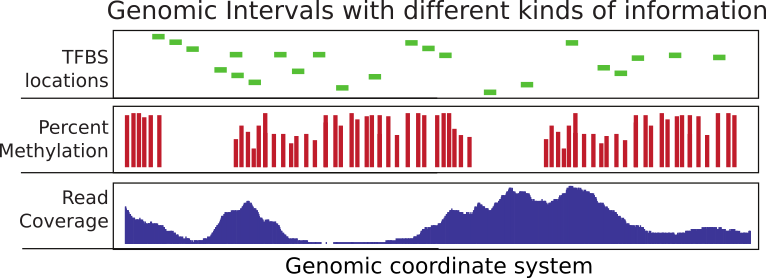
FIGURE 6.1: Summary of genomic intervals with different kinds of information.